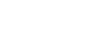Powering Discoveries
Aaron Dubrow
Related Articles
Doctors in a Digital Domain
Advancing medical research across Texas
Texas is home to some of the world’s leading medical research centers. Many are part of The University of Texas System’s 14 academic and health institutions. Researchers at these institutions push the boundaries of medicine, and many use TACC supercomputers to advance their work.
A unique partnership that started over a decade ago, known as The University of Texas Research Cyberinfrastructure, allows any faculty member, researcher, or student at a UT System institution to access TACC resources.
Three areas of medical research that are particularly aided by high performance systems are medical imaging, genomics, and molecular simulation, according to Joe Allen, who leads TACC’s UT System collaborations. “Whether it’s for big data analysis, simulation or AI, Texas medical researchers are making great use of our systems.”
 Corpus callosum, a primary region of interest in traumatic brain injury studies, were reconstructed from Diffusion Tensor Imaging data by UTHealth. Credit: Jenifer Juranek, UT Health
Corpus callosum, a primary region of interest in traumatic brain injury studies, were reconstructed from Diffusion Tensor Imaging data by UTHealth. Credit: Jenifer Juranek, UT Health
Brain Imaging to Assess Treatment Results
Jenifer Juranek, an associate professor of Pediatrics at The University of Texas Health Science Center at Houston (UTHealth), is analyzing brain imaging data from Phase 2 clinical trials of individuals with traumatic brain injuries who have been treated with bone marrow stem cells.
Juranek uses a range of imaging technologies to assess the changes in brain tissue in the days, weeks, and months after an injury, producing huge quantities of data. TACC supercomputers help her compare brain physiology among patients and uncover changes — both for those being treated and those in the control group.
The project, led by Charles S. Cox, Jr., Director of the Pediatric Program in Regenerative Medicine at UTHealth’s McGovern Medical School, is sponsored by the Joint Warfighter Program within the U.S. Army Medical Research Acquisition Activity. Juranek is currently analyzing data from their studies; they hope to move to Phase 3 trials in the coming years.
“If medical research wants to continue to make advances, they need to pair themselves with high performance clusters,” Juranek said. “The information that we're gathering is so massive that in order to analyze it properly, you’ve got to have access to these kinds of resources.”
Identifying Where DNA Breaks and Why
Genomics is another area where access to powerful computers can speed up research or make new lines of inquiry available.
Since 2013, Albino Bacolla, a research scientist at MD Anderson, has been studying the genetic signatures of cancer using TACC supercomputers. One of his projects involves applying advanced computing to identify patterns of mutations in cancer data.
Alterations in the genome are often not random; they tend to occur at specific locations or are related to certain DNA sequences. Using TACC’s Stampede2 and Lonestar5 machines, Bacolla’s team has been investigating whether sequences prone to form non-B DNA conformations — those beyond the classic double-helix — are more likely to experience chromosomal breaks that lead to gross rearrangements in cancer.
The researchers developed search criteria to identify all non-B DNA-forming sequences. By mapping the locations of chromosomal breaks in cancers using the reference human genome, they are capturing the genomic basis of cancer on a broad scale.
“There is an enormous amount of data available in the public domain concerning cancer patients,” Bacolla said. “What is lacking is the ability to analyze and mine the data. HPC resources allow me to query a large number of data to probe its meaning.”
 MD Anderson researchers used TACC’s Stampede2 supercomputer to explore mutations in cancer cells. Credit: Albino Bacolla, MD Anderson
MD Anderson researchers used TACC’s Stampede2 supercomputer to explore mutations in cancer cells. Credit: Albino Bacolla, MD Anderson
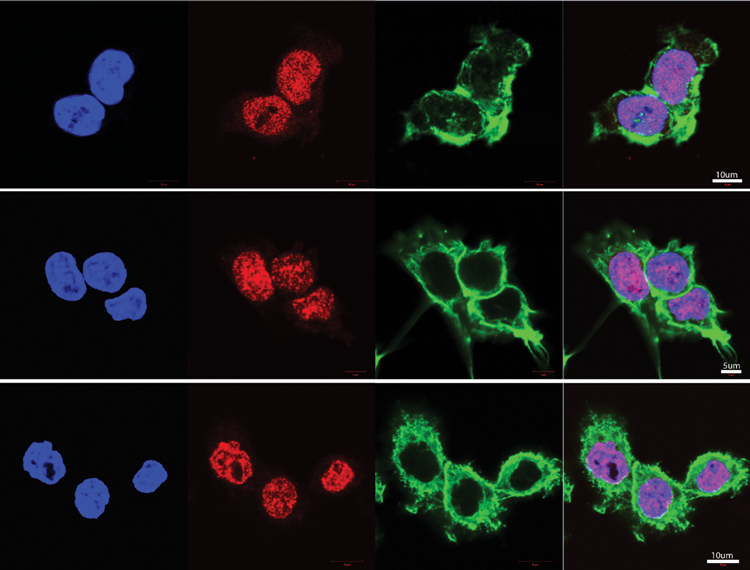
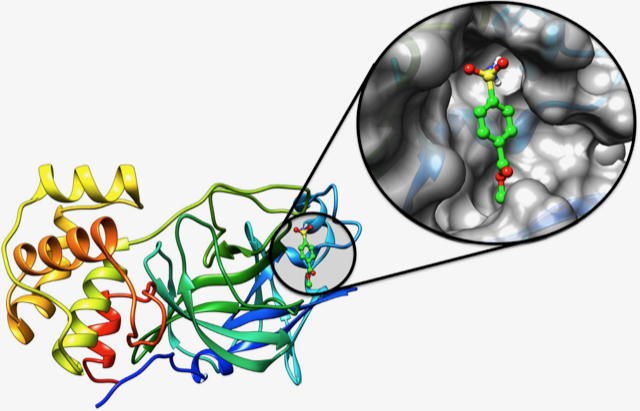 A potential antiviral small molecule docked to the main protease of SARS-CoV-2. Credit: COVID-19 Drug Discovery Consortium
A potential antiviral small molecule docked to the main protease of SARS-CoV-2. Credit: COVID-19 Drug Discovery Consortium
A COVID-19 Collaboration as Big as Texas
In early 2020, an international team of medical researchers — led by UT System scholars — coalesced to rapidly identify drug-like molecules that inhibit SARS-CoV-2 replication.
Known as the COVID-19 Drug Discovery Consortium, by May they had screened 2.6 million small molecules against all the major non-structural proteins of SARS-CoV-2 using TACC supercomputers. They identified 600 small drug molecules that were most likely to be effective and moved quickly to test these against the virus in high containment facilities at Boston University.
After a preliminary analysis, Stan Watowich, professor of Biochemistry & Molecular Biology at UT Medical Branch and team lead, says they have found eight novel compounds that show antiviral activity against SARS-CoV-2.
“Most encouraging, the compounds we identified target different viral proteins and are predicted to disrupt several different and essential SARS-CoV-2 replication pathways,” Watowich said.
Moving forward, they will test whether combinations of these molecules can be effective as treatments for COVID-19.
These three projects represent just a small fraction of the medical research across the state aided by TACC resources. Flu tracking, hearing loss studies, analyses of Alzheimer drug trials — name a disease or medical issue and a UT System researcher is using supercomputers to understand its origin and find a treatment.